CircolettoAn idea that originated from intra-Institute experiences (as was the name actually), and based on Circos, Circoletto visualises sequence similarity from BLAST results (which you can either create through Circoletto, or upload your own). [server] | [citation] | [code] |
 |
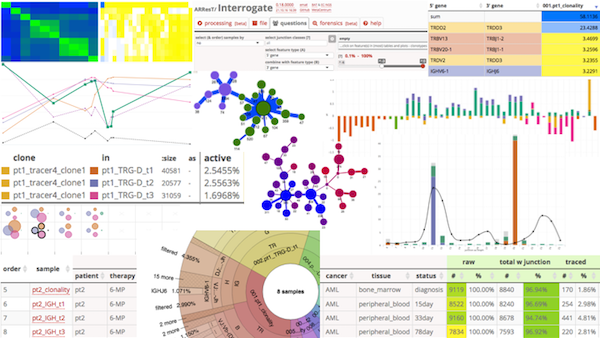
ARResT/InterrogateWeb-based, interactive application for immunoprofiling IG/TR sequence data, both NGS and Sanger. It can process, analyse, organize and filter large amounts of data by numerous criteria and their combinations, based on user-provided arbitrary metadata, and present them in the form of several interactive visualizations. |
 |
ARResT/AssignSubsetsARResT/AssignSubsets was built to robustly assign user-submitted sequences as new members to existing subsets of stereotyped antigen receptor sequences, currently applicable to the 19 major subsets of stereotyped B-cell receptors in chronic lymphocytic leukemia (CLL), through sets of rules captured in a statistical model. That implies that ARResT/AssignSubsets does not discover new subsets. |
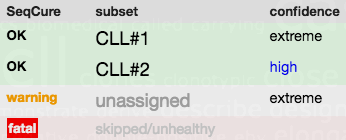 |
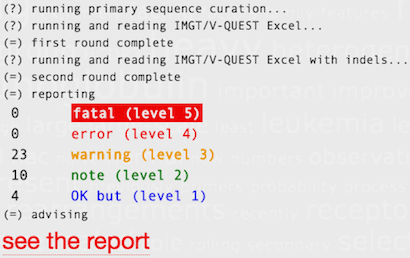
ARResT/SeqCureARResT/SeqCure is able to systematically and consistently capture, describe, and advise on issues with the input antigen receptor sequences based on an easily expandable knowledgebase and set of rules. Quality related issues include: - sequence ambiguities, - problematic length, - sequence and/or ID overlaps. Annotation (with IMGT/V-QUEST) related issues revolve around: - sequences containing insertion/deletions, - sequence with defects (e.g. missing CDR3 anchors), - double productive/single unproductive cases. ARResT/SeqCure consequently allows for higher quality downstream immunogenetic analysis. |
 |
GLASSAssisted and standardized assessment of gene aberrations from Sanger sequence trace data. A collaboration between us, the Medical Genomics Group @ CEITEC MU, the European Research Initiative on CLL / ERIC, and the IgCLL group. |
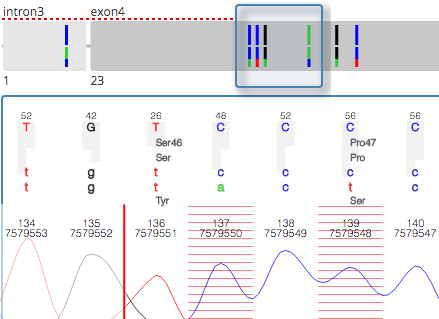 |
